NEUROLOGY PROGRAM
NEUROLOGY - SUBTYPES, TARGETS, AND IN VIVO TRANSLATIONAL EVIDENCE
Neurology programs often fail at the handoff between patient signals and actionable in vivo evidence. Biology is distributed across circuits and systems, symptoms evolve over time, and diagnostic labels often group together distinct biology. When direct, high-fidelity biomarkers are limited, programs need functional and phenotypic readouts that translate.
The gap is decision-grade validation that a mechanism-linked model captures disease-relevant phenotypes and that treatment effects are measurable in the readouts that matter for patient stratification and endpoints.Lunai closes that gap by combining large-scale patient stratification with quantitative in vivo biology. We identify outcome-linked subtypes and progression signals from multimodal data, then translate hypotheses into measurable model-system readouts when in vivo evidence is required. The result is a tighter link between patient subgroup, endpoints, and the evidence used to justify a program direction.

OUR NEUROLOGY ASSETS
Stratification:
Outcome-linked subtypes and biomarkers that support inclusion, enrichment, endpoint strategy, and response assessment.
Target identification:
Progression-linked target hypotheses grounded in trajectories and pathway-level signals.
Validation:
Quantitative in vivo readouts for activity, toxicity, and neurobehavioral, morphological, and seizure-relevant phenotypic correction when functional evidence is required.
PROGRAM EXAMPLE - PARKINSON'S DISEASE SUBTYPING + TARGET PRIORITIZATION
Slow progression signals and biological heterogeneity can obscure therapeutic effects in neurodegeneration, including Parkinson's disease and Alzheimer's disease. The decision need involves distinct subtypes linked to outcomes, paired with biomarker and target hypotheses that support trial design and proof-of-concept.
WHAT WE DID:
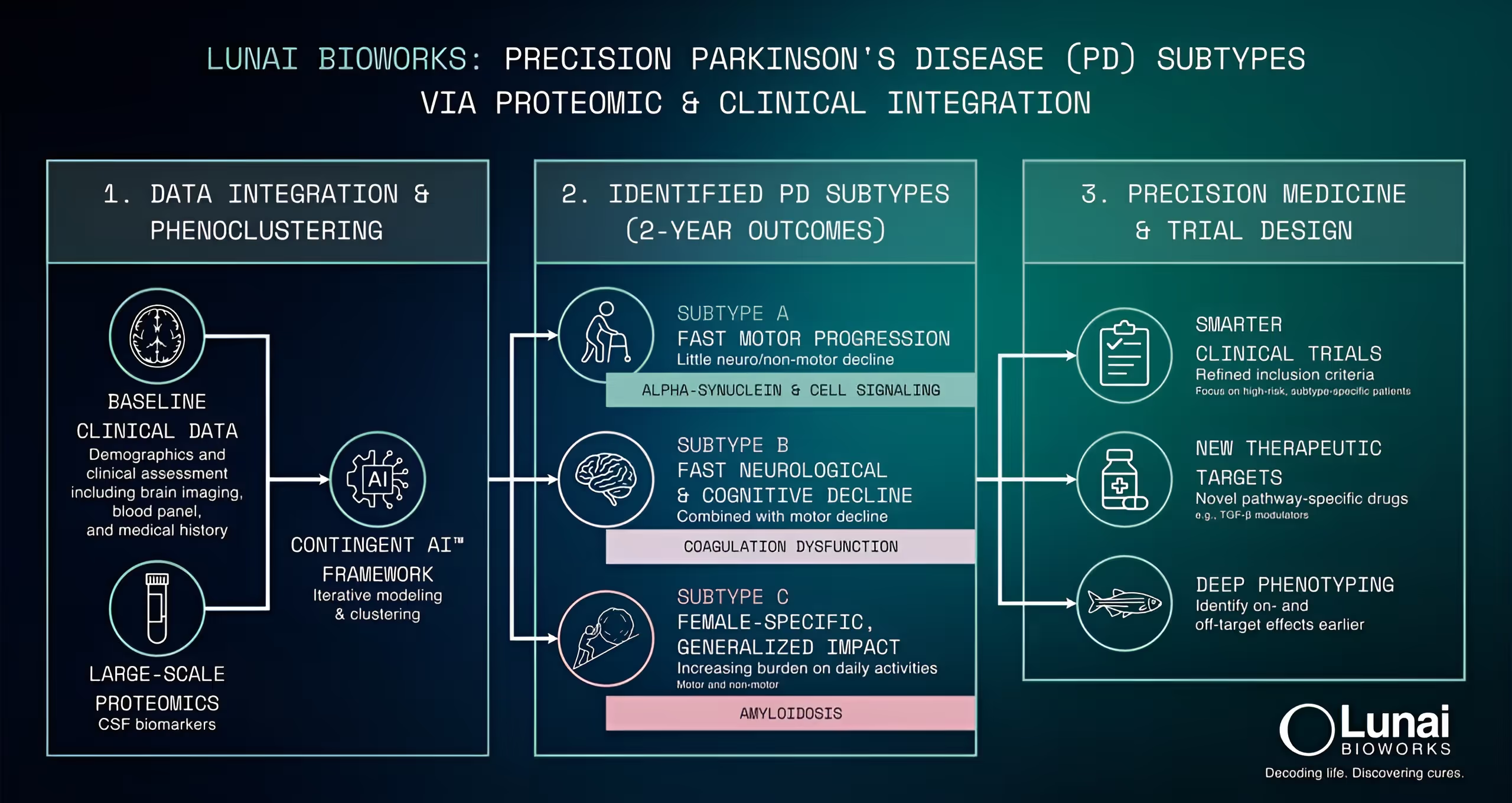
- Cohort integration: We integrated longitudinal clinical phenotypes with proteomics from the Parkinson's Progression Markers Initiative (PPMI), alongside baseline clinical data, analyzing more than 650 participants across 4,500 proteomic probes.
- Subtyping: We performed phenogrouping, calibrated based on the ability to predict outcome labels.
- Cohort integration: We integrated longitudinal clinical phenotypes with proteomics from the Parkinson's Progression Markers Initiative (PPMI), alongside baseline clinical data, analyzing more than 650 participants across 4,500 proteomic probes.

OUTCOMES:
- Three outcome-linked Parkinson's disease subtypes: fast motor progression with limited non-motor involvement alongside cognitive decline; rapid neurological + cognitive decline alongside motor worsening with immune response markers; and a female-enriched subtype with increasing burden on daily activities and an amyloid-associated signature.
- An actionable foundation for inclusion, enrichment, and endpoint strategies aligned to outcome trajectories, with biomarker candidates for baseline stratification, progression monitoring, and assessment of treatment responses.
- A prioritized set of targets and pathways for experimental follow-up and model selection, supported by progression-linked signals.
IMPLICATIONS:
- Faster trial design decisions by decoding heterogeneity into outcome-linked subtypes with clear inclusion and enrichment options.
- Clearer patient-to-biology linkage by tying progression-linked proteins and pathways to biomarker candidates and prioritized targets for follow-up.
- Reduced risk by matching inclusion, enrichment, and endpoints to subtype-level progression signals instead of treating Parkinson's and Alzheimer's as uniform populations.

PROGRAM EXAMPLE - SEIZURE PHENOTYPES + IN VIVO RESPONSE EVIDENCE
This program required fast de-risking of candidate compounds and evidence that genetic models reproduce seizure phenotypes and quantify treatment effects in a way that informs patient response.
WHAT WE DID:
- Compound screening: We screened an uncharacterized compound identified from natural sources across multiple epilepsy models, providing the first in vivo readout of activity and toxicity and comparing its phenotype profile to reference profiles.
- Gene-to-model translation: We used the Phenograph™ to nominate priority genes from clinical phenotypes and defined a translational path to zebrafish genetic models.
- In vivo evidence: We captured behavioral and morphological phenotypes and quantified drug effects on seizure phenotypes across genetic models, assessing phenotypic rescue to support response prediction and stratification strategy.


OUTCOMES:
- Evidence that the compound was not broadly anti-convulsive across the tested epilepsy models, with a profile consistent with a selective serotonin reuptake inhibitor (SSRI) class signature.
- A candidate therapy that reversed seizure phenotypes in two genetic models.
- Zebrafish data that supported a U.S. Food and Drug Administration (FDA) submission, with a Phase 2 trial in drug-resistant epilepsy (DRE) being launched with a patient stratification strategy informed by the genetic models.
IMPLICATIONS:
- Faster go/no-go decisions by screening for activity and toxicity in vivo before committing to deeper development work.
- Reduced translational risk by connecting clinical phenotypes to genetic models with measurable seizure-relevant readouts.
- Decision-grade evidence to support regulatory and clinical next steps, including a stratification strategy informed by genetic models.

KCC2 - RELATED PROGRAM

For a focused view of our phenomics-enabled KCC2 effort, click through to the dedicated page: KCC2 program.

